By going through these Maharashtra State Board 12th Science Biology Notes Chapter 4 Molecular Basis of Inheritance students can recall all the concepts quickly.
Maharashtra State Board 12th Biology Notes Chapter 4 Molecular Basis of Inheritance
The Discovery of DNA-
1. Nuclein:
- Isolated by Friedrich Miescher, in 1869, from the nuclei of pus cells.
- It is an acidic substance with high phosphorus content.
2. There are two types of nucleic acids – DNA (deoxyribonucleic acid) and RNA (ribonucleic : acid).
![]()
The Genetic Material is a DNA-
1. Initially proteins (and not DNA) were ; considered as genetic material because :
- Proteins are large, complex molecules and store information required to govern cell metabolism. Hence, it was assumed that variations found in species were caused by proteins.
- DNA was considered as a small, simple : molecule whose composition varies little ; among species.
- Variations in the DNA molecules are different j than the variation in shape, electrical charge J and function shown by proteins.
2. Various experiments which proved that DNA (and not protein) was the genetic material are as follows :
A. Griffith’s experiments :
- In 1928, Frederick Griffith, carried out an experiment with two strains of bacterium Streptococcus pneumoniae: S-type (Virulent, smooth, pathogenic and encapsulated) and R-type (Non-virulent, rough, non-pathogenic and non-capsulated).
- He observed that on injecting a mixture of heat-killed S bacteria and live R bacteria, the mice died.
- Griffith obtained live S-strain bacteria from the blood of the dead mice.
- Conclusion: Live R-strain bacteria must have picked up something (transforming principle) from the heat-killed S bacterium and got transformed into S-type.
B. Avery, McCarty and MacLeod’s experiment:
- Purified DNA, RNA, proteins, etc. from heat killed cells of S-strain and mixed with R-strain bacteria separately.
- Only DNA was able to transform avirulent R-strain into virulent S-strain.
- When DNA was digested with DNase, there was no transformation.
- Thus, in 1944, they proved that the DNA is a genetic material (transforming principle), but all biologists were not convinced.
C. Hershey-Chase Experiment :
- Hershey and Chase worked with bacteriophages.
- TWo types of bacteriophages were used in the experiment – type one where DNA was labelled with radioactive phosphorus and type two where protein coat was labelled with radioactive sulphur.
- Steps : infection, blending, centrifugation.
- Experiment proved that DNA is the genetic material which enters bacterial cell and not protein.
DNA packaging-
A. Packaging in Prokaryotes :
- E. coli cell size : 2-3μ.
- The nucleoid : Small, circular, highly folded, naked DNA (1100μm long in perimeter, contains about 4.6 million base pairs), nuclear membrane, nucleolus are absent.
- Negatively charged circular DNA (350 μm in diameter).
- Folding / looping : Size reduction to 30 μm in diameter.
- Coiling and supercoiling of each domain : Size reduction to 2μ in diameter.
- Positively charged HU (Histone like DNA binding proteins) proteins : Assist in coiling.
- DNA gyrase and DNA topoisomerase I : Maintain super coiled state.
B. Packaging in Eukaryotes :
- Presence of nuclear membrane, nucleolus and thread-like chromosomes.
- To accommodate long DNA molecule (2.2 m in a typical mammalian cell) in such a small nucleus (10-6 m), it is condensed, coiled and supercoiled.
- R. Kornberg in 1974 reported that DNA is associated with histone and non-histone proteins in the chromosomes.
- Histones: A set of positively charged, basic proteins, rich in basic amino acid residues lysine and arginine.
- Nucleosome: Consists of nucleosome core (two molecules of each of histone proteins viz. H2A, H2B, H3 and H4 forming histone octamer) and negatively charged DNA (146 bps) that wraps around the histone octamer by 1 3/4 turns.
- H1 protein binds the DNA thread where it enters and leaves the nucleosome.
- Adjacent nucleosomes are linked with linker DNA (varies in length from 8 to 114 bp, average length of linker DNA is about 54 bp). [Note : Technically nucleosome includes nucleosome core, DNA wrapped around it (146 bp) and one adjacent linker DNA (54 bp), thus, each nucleosome contains 200 bp of DNA.]
- Packaging involves formation of – Beads on string, Solenoid fibre (looks like coiled telephone wire, 30 nm diameter/300 Å), Chromatin fibre and Chromosome.
- Non-Histone Chromosomal Proteins (NHC) : Additional sets of proteins that contribute to the packaging of chromatin at a higher level.
![]()
C. Heterochromatin and Euchromatin :
1. Heterochromatin :
- This term was proposed by Heitz.
- Genetically (transcriptionally) almost inactive, darkly stained, condensed parts of chromonema/chromosomes observed in eukaryotic cells, during interphase and early prophase.
- Located near centromere, telomeres are also interrelated.
- Heterochromatin is 2 to 3 times richer in DNA than in the euchromatin.
2. Euchromatin : Genetically (transcriptionally) . active, lightly stained, fast replicating regions of chromonema which are in non-condensed state.
DNA Replication-
1. Functions of DNA :
- Regulates and controls all the cellular activities.
- DNA replicates and gets distributed equally to the daughter cells when the cell divides.
- Carrier of genetic information.
- Heterocatalytic function : Directs the synthesis of chemical molecules other than itself. E.g. Synthesis of RNA (transcription), synthesis of protein (Translation), etc.
- Autocatalytic function : Directs the synthesis of DNA itself. E.g. Replication.
- A master molecule of a cell that initiates, guides, regulates and controls the process of protein synthesis.
2. Replication is the process by which DNA duplicates itself and forms two copies that are identical to it.
3. In eukaryotes, replication of DNA occurs once, in the S-phase of interphase.
4. The steps involved in DNA replication :
- Activation of Nucleotides
- Point of Origin or Initiation point
- Unwinding of DNA molecule
- Formation of Y-shaped replicating fork
- Synthesis of new strands
- Leading and Lagging strand
- Formation of daughter DNA molecules
5. Enzymes and proteins involved in DNA replication :
- Phosphorylase
- Helicase
- Single strand binding proteins (SSBP)
- Primase
- DNA polymerase
- DNA ligase
- Super-helix relaxing enzyme
- DNA gyrase (Topoisomerase)
6. Semiconservative replication : In each daughter DNA molecule, one strand is parental and the other one is newly synthesized. Thus, 50% mother DNA is conserved.
7. Experimental confirmation of semiconservative DNA replication : Given by Matthew Meselson and Franklin Stahl (1958). They used light and heavy isotopes of nitrogen and equilibrium-density-gradient-centrifugation technique.
![]()
Protein synthesis-
1. Central Dogma :
(1) Postulated by F.H.C. Crick in 1958.
![]()
Transcription Translation
(2) In eukaryotes, DNA transcription takes place in nucleus and translation occurs in cytoplasm. In prokaryotes, both the processes occur in cytoplasm.
(3) Central dogma in retroviruses : Temin (1970) and Baltimore (1970)

2. Transcription :
(1) Transcription is the process of copying of genetic information from one (template) strand of DNA into a complementary single stranded RNA transcript.
(2) Occurs in the nucleus during Gx and G2 phases of cell cycle.
(3) Catalyzed by RNA polymerase :
(i) Prokaryotes : One type of RNA polymerase.
(ii) Eukaryotes :
- RNA polymerase-I : Transcription of r-RNA.
- RNA polymerase-II : Transcription of m-RNA and heterogeneous nuclear- RNA (or hnRNA).
- RNA polymerase-III : Transcription of t-RNA and small nuclear-RNA (snRNA).
(4) Transcription Unit: Transcription unit (Each transcribed segment of DNA) consists of the promoter, the structural gene and the terminator.
(i) The promoter :
- Located towards 5′ end of structural gene, i.e. upstream.
- Provides binding site for enzyme RNA polymerase.
(ii) Structural genes :
- Template strand : DNA strand having 3′->5′ polarity.
- Sense strand : The other strand of DNA having 5′->3′ polarity.
(iii) The terminator :
- Located at 3′ end of coding strand, i.e. downstream.
- Defines the end of the transcription process.
(5) Three stages of transcription : Initiation, Elongation and Termination.
(6) Transcription unit and the gene :
- Gene : The DNA sequence coding for m-RNA/t-RNA or r-RNA.
- Cistron : A segment of DNA coding for a polypeptide.
- Monocistronic gene : A single structural gene in transcription unit.
- Polycistronic gene : One transcription unit having a set of various structural genes.
- Interrupted genes (Split genes) :
Structural genes with both exons and introns).- Exons : The coding sequences or express sequences.
- Introns : The non-coding sequences.
- Only exons appear in processed m-RNA in eukaryotes.
- In bacteria. m-RNA does not require any processing because it has no introns.
(7) Processing of hnRNA :
- Primary transcript or hnRNA is non¬functional and contains both exons and introns.
- Processing of hnRNA results in functional m-RNA.
- hnRNA undergoes capping, tailing and splicing.
- Capping : Methylated guanosine tri¬phosphate is added to 5′ end of hnRNA.
- Tailing : Polyadenylation take place at 3′ end.
- Splicing : Removal of introns.
- DNA ligase joins exons in a definite sequence (order).
- The fully processed hnRNA is called m-RNA.
![]()
(8) Genetic Code :
- Yanofski and Sarabhai (1964) : Provided evidence that DNA carries the information for the protein synthesis as the sequence of nucleotides.
- F.H.C. Crick : According to Crick the information for protein synthesis is stored in the form of coded language (cryptogram) called genetic code. He gave evidence for the triplet nature of genetic code, by using “frame-shift mutation”.
- G. Gamow (1954) : Suggested that codon is a sequence of three consecutive nucleotides on m-RNA.
- Cracking of genetic code : M. Nirenberg, Matthaei, Ochoa and Har Gobind Khorana : Deciphered complete genetic code by using artificial m-RNA templates (homopolymers and copolymers) and cell free system of protein synthesis. Synthesized artificial poly-U RNA.
(a) M. Nirenberg and Matthaei : Synthesis of poly-U, m-RNA and polypeptide that consisted of only phenylalanine.
(b) Har Gobind Khorana : Devised a technique for synthesis of artificial m-RNA with repeated sequences of known nucleotides.
(c) Severo Ochoa : The enzyme polynucleotide phosphorylase polymerizes RNA with defined sequences in a template-independent manner (i.e. enzymatic synthesis of RNA).
(9) Replication and transcription are based on complementarity principle.
(10) During translation, complementarity principle is not applicable as, genetic information is transferred from a polymer of nucleotides to a polymer of amino acids.
(11) Characteristics of Genetic code :
- Triplet code : Codon (sequence of three consecutive nucleotides), specifies one particular amino acid.
- Polarity : Genetic code is always read in 5′ -» 3′ direction.
- Non-overlapping code : Each single nucleotide is a part of only one codon.
- Commaless : There is no gap between successive codons.
- Degeneracy of genetic code : Two or more codons can specify the same amino acid. E.g. Cysteine has two codons, while isoleucin has three codons.
Degeneracy of the code is explained by Wobble hypothesis. - Universal code : In all living organisms the specific codon specifies same amino acid. E.g. codon AUG always specifies amino acid methionine.
- Non-ambiguous code : Each codon specifies a particular amino acid.
- Initiation codon : AUG, Codes for amino acid methionine.
- Termination codons : UAA, UAG and UGA : They do not code for any amino acid. They stop the process of elongation of polypeptide chain.
- Codon : A triplet of nucleotides present on DNA that codes for specific amino acid. E.g. AUG is codon.
- Anticodon : Triplet of nucleotides present on the anticodon loop of t-RNA, which is complementary to codon on m-RNA.
- Wobble hypothesis : In codon-anticodon pairing the third base may not be complementary.
(12) Mutation recombination :
(i) Mutation : A sudden heritable change in the DNA sequence that results in the change of genotype.
(ii) Mutation and recombination is raw material for evolution as it generates variations.
(iii) Types of mutations :
- Chromosomal mutations : Loss (deletion) or gain (insertion/ duplication) of a segment of DNA results in alteration in the chromosome.
- Point mutations : Involve change in a single base pair of DNA. E.g. mutation that results in Sickle-cell anaemia.
- Deletion or insertion of base pairs of DNA : It causes frame-shift mutations or deletion mutation.
(13) t-RNA – the adapter molecule :
- t-RNA can read the codon and also can bind with the amino acid. So t-RNA is considered as an adapter molecule.
- Cloverleaf structure (2 dimensional) of t-RNA has 3 loops and 4 arms.
- For every amino acid, there is specific t-RNA.
- Initiator t-RNA is specific for methionine.
- There are no t-RNAs for stop codons.
- In the actual structure, the t-RNA molecule looks like inverted L (3 dimensional structure).
3. Translation – protein synthesis :
- Translation is the process in which sequence of codons on m-RNA is decoded and accordingly amino acids are added in specific sequence to form a polypeptide on ribosomes.
- It requires 20 different amino acids, m-RNA, t-RNA, ribosomes, ATP Mg++ ions, enzymes, elongation, translocation and release factors.
- Translation involves
- Activation of amino acids and formation of AA-t-RNA complex.
- Formation of polypeptide chain : initiation, elongation, termination.
![]()
Regulation of gene expression-
In eukaryotes, the regulation of gene expression can be at different levels like
- Transcriptional level (formation of primary transcript).
- Processing level (regulation of splicing).
- Transport of m-RNA from nucleus to the cytoplasm.
- Translational level.
Operon concept-
1. A transcriptional control mechanism of gene regulation.
2. Francois Jacob and Jacques Monod (1961) : Explained that metabolic pathways are regulated as a unit.
3. Lac operon of E. coli :
(1) An inducible operon.
(2) The operon is switched on by a chemical inducer-lactose present in the medium.
(3) Lac operon consists of the following components :
- Regulator gene (repressor gene)
- Promoter gene
- Operator gene
- Structural genes : z (codes for galactosidase), y (codes for β-galactoside permease) and a (codes for transacetylase). These enzymes are involved in lactose metabolism.
4. Inducer is not a component of operon.
Genomics-
1. The term Genome was introduced by H. Winkler in 1920.
2. Genome : The total genetic constitution of an organism or a complete copy of genetic information (DNA) or one complete set of chromosomes (monoploid or haploid) of an organism.
3. Genomics (term coined by T.H. Roderick in 1986) : The study of genomes through analysis, sequencing and mapping of genes along with the study of their functions.
4. Two types of Genomics :
- Structural genomics : Involves mapping, sequencing and analysis of genome.
- Functional genomics : Involves the study of functions of all gene sequences and their expressions in organisms.
5. Application of genomics :
- Structural and functional genomics are used in the improvement of crop plant, human health and livestock.
- The knowledge and understanding acquired by genomics research can be applied in medicine, biotechnology and social sciences.
- It helps in the treatment of genetic disorders through gene therapy.
- To develop transgenic crops having more desirable characters.
- Genetic markers have applications in forensic analysis.
- Genomics can lead to introduce new gene in microbes to produce enzymes, therapeutic proteins and even biofuels.
Human Genome Project-
1. Initiated in 1990 under the International administration of the Human Genome Organization (HUGO).
2. Coordinated by the US Department of Energy and National Institute of Health, some universities across the United States and various international partners.
3. Started in 1990 and completed in 2003.
4. Aims of HGP :
- Mapping the entire human genome at the level • of nucleotide sequences.
- To store the information collected from the, project in databases.
- To develop tools and techniques for analysis of the data.
- Transfer of the related technologies to the * private sectors, such as industries.
- Taking care of the legal, ethical and social issues which may arise from project.
- To provide complete and accurate sequence of the 3 billion DNA base pairs that make up the human genome.
- To find out the estimated number of human ; genes. Now about 33,000 genes have been : estimated to be present in humans.
- To sequence the genomes of several other organisms such as bacteria e.g. E.coli, Caenorhabditis elegans, Saccharomyces cerevisiae, Drosophil, rice, Arabidopsis), Mus musculus etc.
5. HGP was closely associated with rapid development of Bioinformatics.
6. Significance :
- Increased knowledge about the functions of genes and proteins.
- A major impact in the fields like Medicine, Biotechnology and the Life sciences.
- Increased understanding of gene structure and function in other species, such studies will enhance understanding of human evolution.
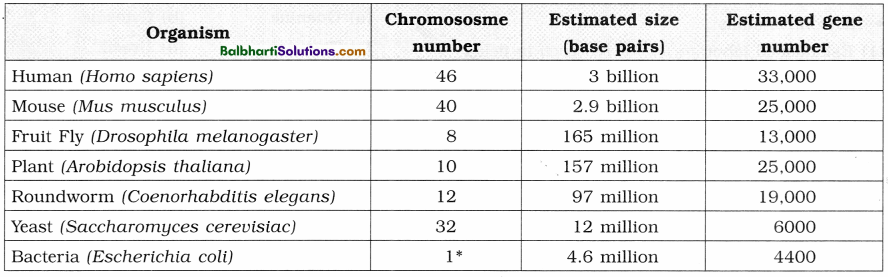
7. Comparative genome sizes of humans and other model organisms.
![]()
DNA Fingerprinting-
1. Every individual has its unique genetic make- up, called its Fingerprint.
2. Reasons for Uniqueness of fingerprint :
- Recombination of paternal and maternal genes, because of which we differ from our parents.
- Infrequent mutations that occur during gamete J formation (cell division).
3. The DNA profiling or DNA fingerprinting j technique (developed by Dr. Alec Jeffreys in 1984) identifies a person with the help of DNA restriction analysis.
4. It is based on identification of nucleotide sequence present in DNA.
- About 99.9% of nucleotide sequence in all persons, is same.
- Variable Number of Tandem Repeats (VNTRs) : Unusual sequences of 20 – 100 base pairs, which are repeated several times.
- As the length of the regions having VNTRs is different in each individual, they are the key factor in DNA profiling.
5. Steps involved in DNA finger printing :
- Isolation of DNA
- Restriction digestion
- Gel electrophoresis
- Southern blotting
- Selection of DNA probe
- Hybridization
- Photography
6. Application of DNA fingerprinting :
- Used in forensic sciences to solve rape and murder cases.
- Finds out the biological father or mother or both, of the child, in case of disputed parentage.
- Used in pedigree analysis in cats, dogs, horses and humans.
Know the scientist :
Dr. Lalji Singh (1947-2017) :
- Father of DNA fingerprinting in India.
- A unique segment obtained from Y chromosome of female banded krait snake (banded krait minor – BKM-DNA) was used by him to develop probe for DNA fingerprinting.
His contributions :
- Established laboratories for research in fields like genetics, population biology, structural biology and transgenics.
- Established centre for DNA Fingerprinting and Diagnostics (CDFD) for all species and several diseases.
- Founded laboratory for Conservation of Endangered Species (LaCONES).
- Applied of DNA fingerprinting technology for wildlife conservation, forensics, evolution and phylogenetic research.